
Analysis of the gut microbiota does not show inherent differences in bacterial composition between wild-type and genetically modified animals. Consistent with studies of the effects of human ATG16L1 polymorphisms, models exhibit morphological abnormalities in both Paneth and goblet cells, but do not develop spontaneous intestinal permeability or inflammatory bowel disease. These are the first reported rat strains with alterations of the Atg16l1 gene. Using CRISPR-Cas9 genome editing technology, we developed a knock-in rat model for the human ATG16L1 T300A CD risk polymorphism, as well as a knock-out rat model to evaluate the role of Atg16l1 in autophagy as well as its potential effect on CD susceptibility. We identified four alternative splice variants with tissue-specific expression. To generate novel rat strains carrying genetic alterations in the rat Atg16l1 gene, we first characterized the wild-type rat gene. The mechanism behind this increased susceptibility is still being elucidated, however, the amino acid change caused by this point mutation results in increased ATG16L1 protein sensitivity to caspase 3-mediated cleavage. A single, nonsynonymous adenine to guanine polymorphism resulting in a threonine to alanine amino acid substitution (T300A) directly preceded by a caspase cleavage site (DxxD) causes an increased susceptibility to Crohn’s disease (CD) in humans. *Parts of this article were derived from the SnapGene website.ATG16L1 is a ubiquitous autophagy gene responsible, in part, for formation of the double-membrane bound autophagosome that delivers unwanted cellular debris and intracellular pathogens to the lysosome for degradation. Contact the HSLS Molecular Biology Information Service with any additional questions. The FAQs also provide support related to files, features, primers, and more. The user guide offers extensive lessons ranging from searching and colors, to enzymes and restriction cloning & linear ligation. Register for SnapGene and check out the tutorial videos.
YOUTUBE SNAPGENE INFUSION CLONING SOFTWARE
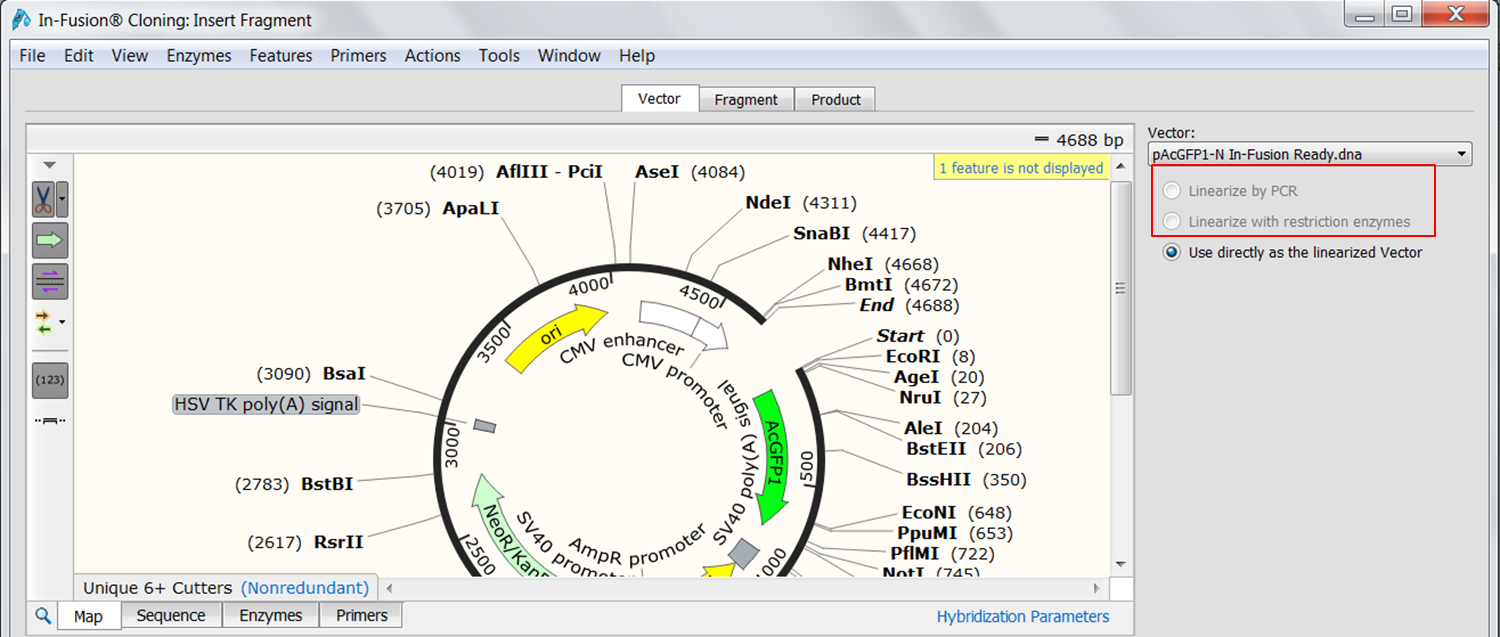
In-Fusion Cloning: SnapGene is the first software to simulate Clontech’s versatile method for creating seamless gene fusions.
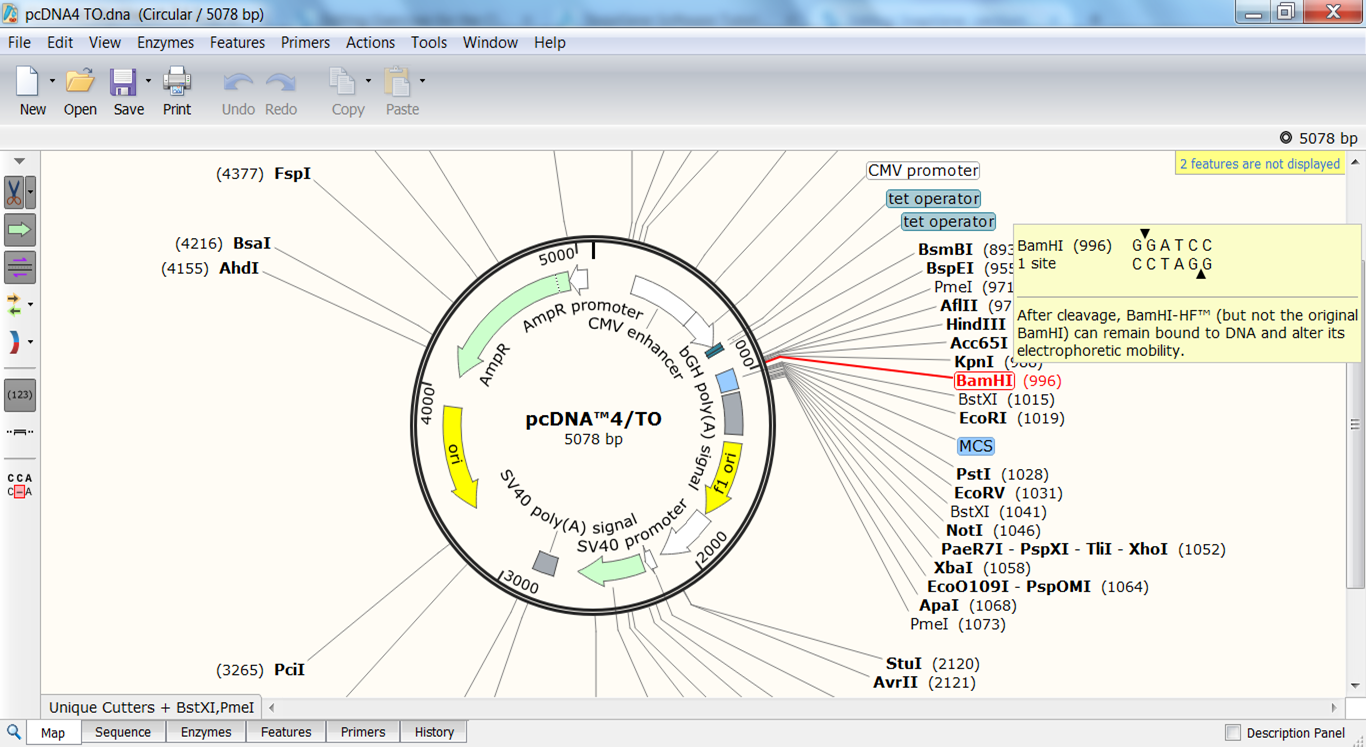
Here are just a few of the things SnapGene can do: The HSLS Molecular Biology Information Service now provides access to this popular molecular biology software, making it that much easier for Pitt researchers to easily perform in silico DNA analysis, molecular cloning, and PCR. You can do all of this and more with SnapGene. share annotated sequence files with other researchers?.automatically record the steps in a cloning project?.visualize ORFs, reading frames, and primer binding sites?.easily plan and simulate your DNA manipulations?.


 0 kommentar(er)
0 kommentar(er)
